Molecular basis for maternal inheritance of human mitochondrial DNA
- PMID: 37723262
- PMCID: PMC10763495
- DOI: 10.1038/s41588-023-01505-9
Molecular basis for maternal inheritance of human mitochondrial DNA
Abstract
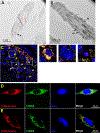
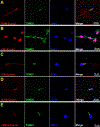
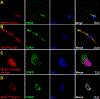
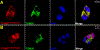
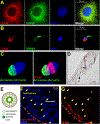
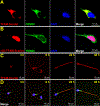
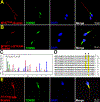
Uniparental inheritance of mitochondrial DNA (mtDNA) is an evolutionary trait found in nearly all eukaryotes. In many species, including humans, the sperm mitochondria are introduced to the oocyte during fertilization1,2. The mechanisms hypothesized to prevent paternal mtDNA transmission include ubiquitination of the sperm mitochondria and mitophagy3,4. However, the causative mechanisms of paternal mtDNA elimination have not been defined5,6. We found that mitochondria in human spermatozoa are devoid of intact mtDNA and lack mitochondrial transcription factor A (TFAM)-the major nucleoid protein required to protect, maintain and transcribe mtDNA. During spermatogenesis, sperm cells express an isoform of TFAM, which retains the mitochondrial presequence, ordinarily removed upon mitochondrial import. Phosphorylation of this presequence prevents mitochondrial import and directs TFAM to the spermatozoon nucleus. TFAM relocalization from the mitochondria of spermatogonia to the spermatozoa nucleus directly correlates with the elimination of mtDNA, thereby explaining maternal inheritance in this species.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests:
Authors declare that they have no competing interests.
Figures







Similar articles
-
Mitochondrial Deoxyribonucleic Acid (mtDNA), Maternal Inheritance, and Their Role in the Development of Cancers: A Scoping Review.Cureus. 2023 Jun 1;15(6):e39812. doi: 10.7759/cureus.39812. eCollection 2023 Jun. Cureus. 2023. PMID: 37397663 Free PMC article.
-
Mitochondrial DNA removal is essential for sperm development and activity.EMBO J. 2025 Mar;44(6):1749-1773. doi: 10.1038/s44318-025-00377-5. Epub 2025 Feb 11. EMBO J. 2025. PMID: 39934414 Free PMC article.
-
Poldip2 promotes mtDNA elimination during Drosophila spermatogenesis to ensure maternal inheritance.EMBO J. 2025 Mar;44(6):1724-1748. doi: 10.1038/s44318-025-00378-4. Epub 2025 Feb 11. EMBO J. 2025. PMID: 39934413 Free PMC article.
-
The dysregulation of mitochondrial homeostasis-related genes could be involved in the decrease of mtDNA copy number in systemic lupus erythematosus patients.Immunol Res. 2024 Dec;72(6):1384-1392. doi: 10.1007/s12026-024-09535-z. Epub 2024 Sep 4. Immunol Res. 2024. PMID: 39230799 Free PMC article.
-
A Scoping Review Investigating the "Gene-Dosage Theory" of Mitochondrial DNA in the Healthy Skeletal Muscle.Int J Mol Sci. 2023 May 2;24(9):8154. doi: 10.3390/ijms24098154. Int J Mol Sci. 2023. PMID: 37175862 Free PMC article.
Cited by
-
Evolution and maintenance of mtDNA gene content across eukaryotes.Biochem J. 2024 Aug 7;481(15):1015-1042. doi: 10.1042/BCJ20230415. Biochem J. 2024. PMID: 39101615 Free PMC article. Review.
-
MtDNA deletions and aging.Front Aging. 2024 Feb 15;5:1359638. doi: 10.3389/fragi.2024.1359638. eCollection 2024. Front Aging. 2024. PMID: 38425363 Free PMC article. Review.
-
Familial coaggregation and shared genetic influence between major depressive disorder and gynecological diseases.Eur J Epidemiol. 2024 Oct;39(10):1161-1170. doi: 10.1007/s10654-024-01166-w. Epub 2024 Nov 4. Eur J Epidemiol. 2024. PMID: 39495462
-
Selectively advantageous instability in biotic and pre-biotic systems and implications for evolution and aging.Front Aging. 2024 May 16;5:1376060. doi: 10.3389/fragi.2024.1376060. eCollection 2024. Front Aging. 2024. PMID: 38818026 Free PMC article. Review.
-
Spatial detection of mitochondrial DNA and RNA in tissues.Front Cell Dev Biol. 2024 May 14;12:1346778. doi: 10.3389/fcell.2024.1346778. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38808224 Free PMC article.
References
-
- Wallace DC Why do we still have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annu Rev Biochem 76, 781–821 (2007). - PubMed
-
- Sutovsky P et al. Ubiquitinated sperm mitochondria, selective proteolysis, and the regulation of mitochondrial inheritance in mammalian embryos. Biol Reprod 63, 582–90 (2000). - PubMed
-
- Sato M & Sato K Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science 334, 1141–4 (2011). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

